Covid-19: The Guide Book
Background:
Over the third Lockdown we started working on producing 3D printed models of the Covid-19 virus. Our latest goal was to produce a low cost paper model, in the style of an origami instruction, this to be used as an educational tool for younger students. This guide has been written to be read alongside all of our models to increase understand and answer some common questions about the virus.
Introduction:
A lesson we are all learning together is that pandemics are scary. No one alive today can remember a situation like this, and no one knowns quite how to bring it to an end. Covid-19 is a disease that spreads invisibly in the air; only 18 months ago it didn’t exist in humans [1]. Although most people, especially younger ones, who catch it suffer a flu-like illness, too many fall seriously ill and several million have died worldwide [2-3]. However, there have been many pandemics before [1], and we now know a great deal about the virus that causes Covid-19. Critically, we now have have multiple vaccines. Here in this booklet, we look at the background and molecular biology of the SARS-CoV-2 virus that causes Covid-19.
Winter – 2019
The latter half of 2019 saw a coronavirus managed to make the jump from infecting bats to infecting humans. This quickly spread and became an outbreak centred around Wuhan in China [1]. Early mutations probably assisted the virus to infection humans making it more contagious [4]. Partly thanks to the mass availability of international air travel, clusters of undetected cases were occurring around the world by the last days of the year [5].
First Half of 2020
By the time scientists and medics understood that the disease was serious enough to make a lot of people ill and over stretch healthcare services – and managed to convince governments of this – it was already starting to circulate around the world. Covid-19 had low rates of hospitalisation and fatality, but when applied to large populations, hospitals quickly become overstretched. This happened first in Italy and then briefly in New York City [6].
Second Half of 2020
The rest of 2020 was punctuated with various lockdowns and restrictions across the countries of the world. As infection levels rose and fell restrictions increased or were loosed with the aim to control the R0 rate of the Covid-19 pandemic. The R0 rate is the average number of people infected by a someone suffering an infectious disease [11]. When the R0 rate for covid-19 is less than 1 the number of cases is falling.
First Half of 2021
2021 has seen an impressive vaccination program in the UK with around 60% of adults fully vaccinated by late June [12]. This together with the measures to supress infectivity, has kept the R0 rate in check. Mutations which the Alpha and Delta variants possess, give the virus a better chance at spreading to the unvaccinated [13]. As countries like Britain reach herd immunity, the new threat will be from variants which can infect vaccinated people.
Dangers of Covid-19:
The average risk of having to go into hospital with Covid-19 is often estimated to be around 10% [7-9], the case fatality rate for all people worldwide is around 2% [10]. The risks increase with age, the pre-delta variant data suggests for those falling ill with Covid-19 in their twenties and below, fewer than 1% were admitted to hospital [8]. However, even if only 10% of a population of 60 million need to be admitted to hospital, demand for hospital beds can quickly out-strip supply. Preserving healthcare services, is the key reason for the numerous lockdowns.
Infection Cycle:
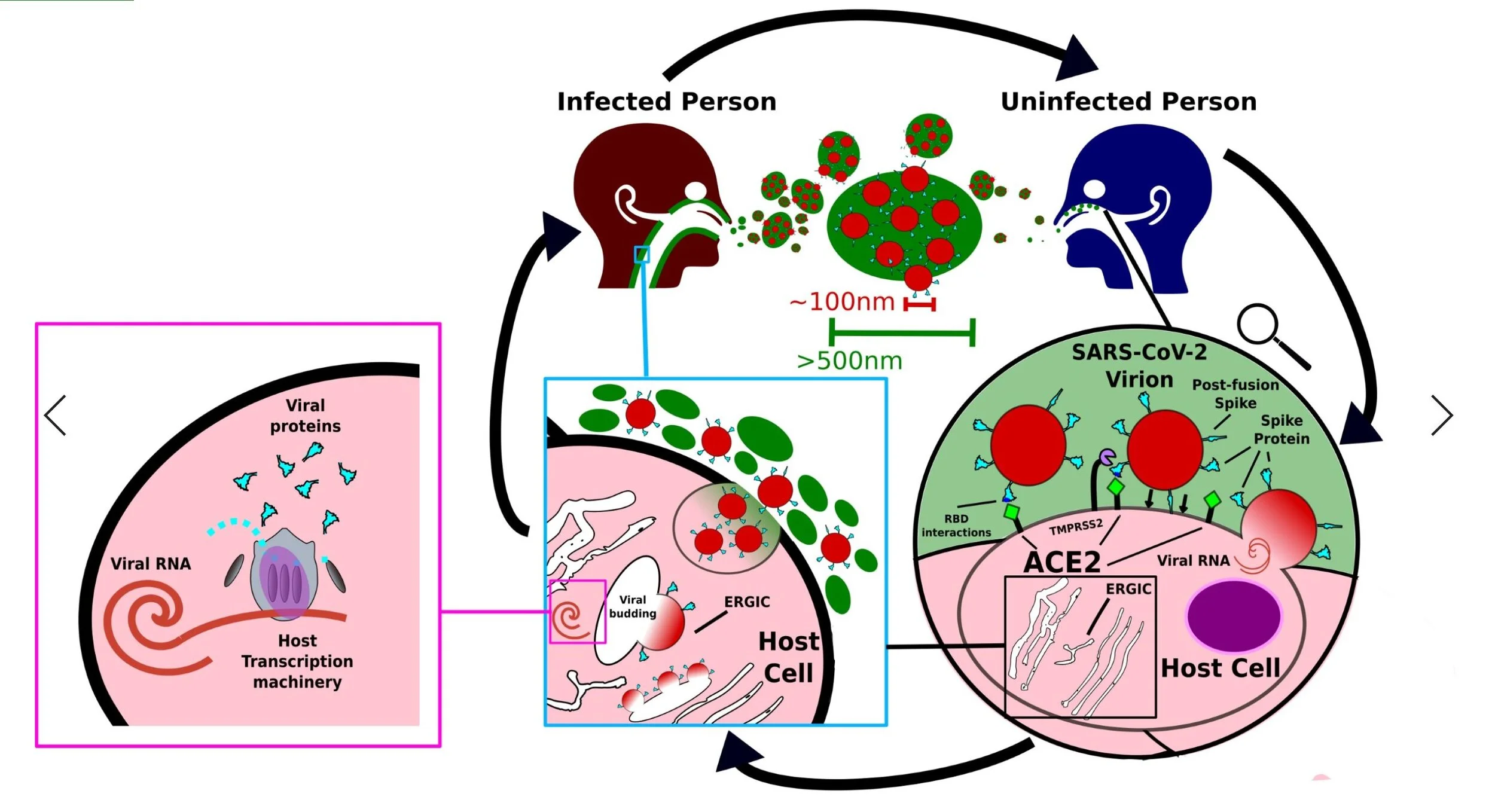
Covid-19 spreads through aerosols emitted from an infected person [14]. Virions, single mature viral particles are suspended in tiny droplets of mucus about 0.5-1.2nm in diameter [15]. Larger particles of 5000nm tend to fall to the ground within 1 meter [16]. Infectious particles can be disseminated through coughing, sneezing, even talking. The Virus can also survive on contaminated surfaces for up to a week [17].
When a Covid-19 virion comes into proximity with an exposed cell usually in the nasal cavity [18], the Spike proteins which dangle off the outside of the virus can recognise and attach to human protein called Angiotensin-converting enzyme 2 (ACE2), this is found hanging off the outside of the human cells lining the nose and throat [19]. The virus uses this interaction between its Spike protein and ACE2 to fuse with or enter the cell, thus allowing its genetic material (viral RNA) to be deposited.
The viral RNA is taken by the cell and read – transcribed into proteins – in the same way native human DNA and RNA is processed [20]. In this way the virus hijacks the human cells into creating viral proteins, these are assembled in a specific part of the cell known as the Endoplasmic-Reticulum-Golgi Intermediate Compartment (ERGIC for short) [21]. Once a sufficient amount of Covid-19 proteins have been made and included in the ERGIC membranes, viral budding occurs [21]. These brand-new viruses are trafficked to the outside of the cell where they are excreted into the mucus and saliva to infect more adjacent cells or emitted into the outside world to infect other hosts.
Biology of Covid-19:
The roughly spherical 100nm outer shell of the SARS-CoV-2 virion – the viral envelope – is formed of a lipid bilayer [22-23] – this is why detergents and alcohol kill the virus. It derives this membrane from the host’s own cellular membranes specifically that of the intracellular membranes of the ERGIC [24-25].
The viral envelope contains the RNA genome to code for the 28 or so proteins of SARS-CoV-2 [26-27]. Most of these proteins only exist in infected cells during viral replication. Only the Spike (S), Membrane (M), and Envelope (E) proteins are found on the virion’s surface. With the Nucleocapsid protein found inside the viral envelope helping to secure the RNA and stop tangling [28].
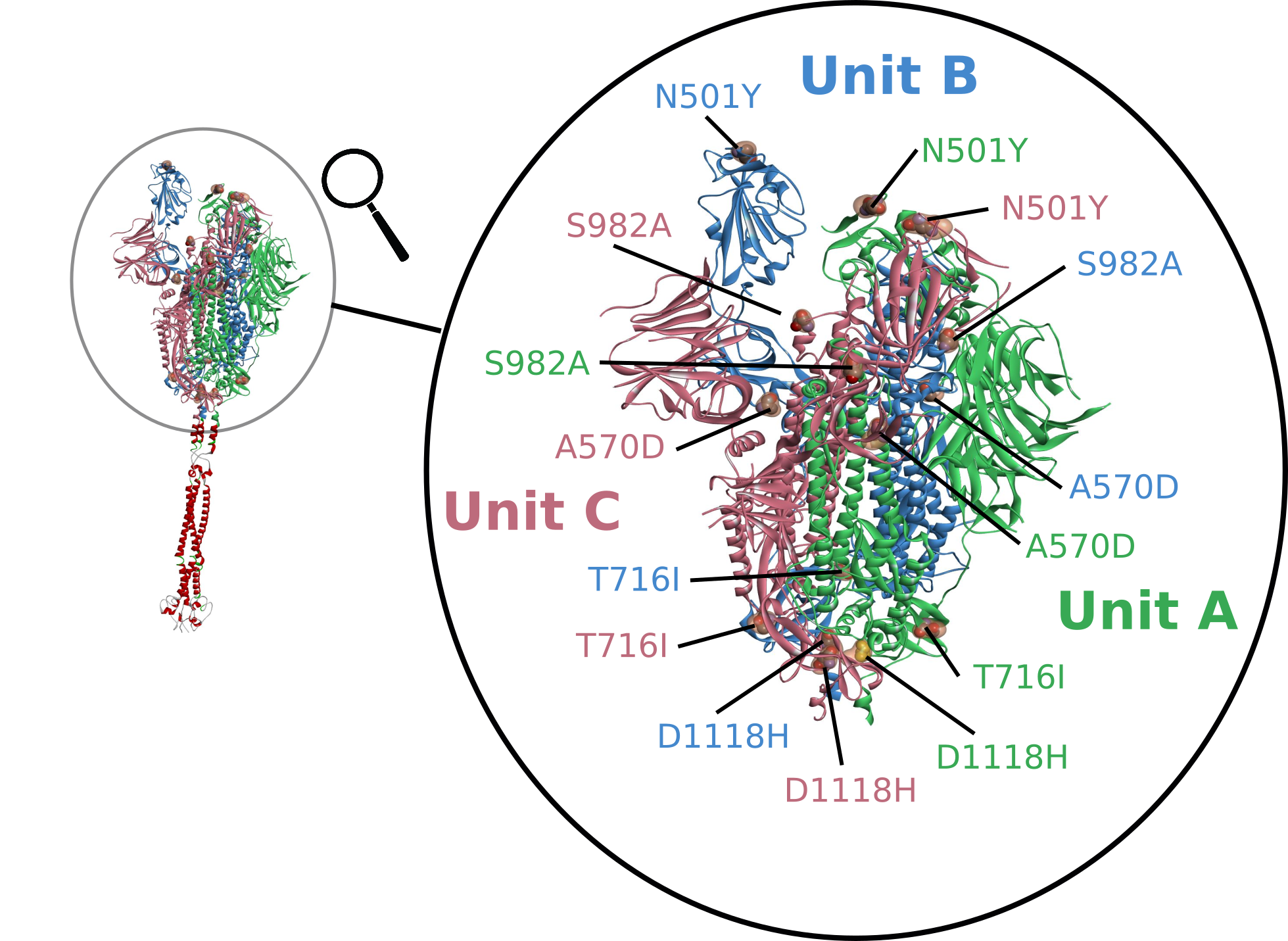
Mutations which alter the proteins of SARS-CoV-2 especially ones which increase infectivity, have occurred throughout the pandemic [29]. The most concerning changes have affected the Spike protein paticularly around the receptor-binding domain (RBD). This part of the virus is the key that unlocks the door to infecting our cells. The Spike protein is made from three identical units each built of around 1200 amino acids [30]. The Kent (Alpha) variant has a total of 17 mutations of these 2 deletions and 6 changes affect the Spike protein, we show the mutations N501Y, A570D, T716I, S982A, and D1118H below [31]. The Alpha variant has helped cause new waves of infection, being 50-75% more contagious than the original strain [32]. These tiny molecular differences found in the variants are hardly noticeable on protein models yet can have devastating effects of transmissibility, fatality, even vaccine resistance [33,34,35].
The Spike protein has specific regions – receptor binding domains – which interacts with the human membrane protein ACE2 and facilitates the virion entering the cell [36]. Thanks to electron micrograph studies looking at and analysing images of actual SARS-CoV-2 virions we known each individual virus has around 20-30 Spike proteins hanging off it [37].
These have 4 key shapes. The first is the closed conformation where the receptor binding domains are hidden – like a cover on a paddle lock. The second is the open conformation where one receptor binding domain is ready to interact with ACE2. The next is double open conformation where two receptor binding domains are exposed. The final one is the post-fusion form; this has a smaller structure, as part of the protein has been cut away [38]. The Spike protein takes this form after it has interacted with ACE2 and another protein known as TMPRSS2 – this conformation can occur spontaneously without any input from ACE2 [19]. Any mature Covid-19 virion is likely to have a mixture of all the forms of the S protein, the most common Spike conformation being the open one [37,39].
The Membrane protein is probably the most numerous with 400-2000 units per virion [40]. The protein is inset in the lipid bilayer, likely serving to anchor the Spike and Nucleocapsid and helping maintain the envelope’s structural integrity [41]. Of the key proteins this one is the least understood, the structure has been modelled but not yet directly observed.
Finally, the Envelope protein takes the form of a tube, the small hole in the centre probably allowing ions to transverse the lipid bilayer [42]. Only a small number are included in the mature virion, it seems likely this protein serves to aid viral budding and may not have an important role thereafter [43].
Vaccination:
The two types of vaccine given in the UK both introduced the RNA code for the Covid-19 Spike protein [44[. The AstraZeneca-Oxford (AZ) vaccine uses an inactive virus which can enter cells but not replicated. It uses an altered chimpanzee adenovirus implanted with the RNA code for the Spike protein [45]. The human cell takes the RNA code and translates it into viral Spike proteins. The body’s immune response identifies the vaccine as part of pathogen and learns to produce antibodies that fit and then block the introduced Spike protein.
The Pfizer-BioNTech vaccine uses an artificial lipid-nanoparticle to encase the RNA code of the Spike protein [46]. The lipid-nanoparticle introduces the RNA to cells around the body, this is read and made into Spike proteins which similar to the AZ vaccine trigger the body to produce suitable antibodies.
In both cases the body learns how to create proteins that can recognise the SARS-CoV-2 Spike protein. Although both vaccines have been approved on an emergency basis, the processes behind how they are made and work is well understood each vaccines safety and efficiency carefully studied [47-50]. Vaccination is a medical procedure, the decision to be carefully thought through. However, to dismiss the one in a hundred risk of mortality posed by Covid-19 and stress at the 1 in a million-risk associated with the vaccine is plainly irrational.
Summary:
From the simple background, set out here, we can understand a lot. Firstly, detergents and alcohol gels will degrade and destroy the virus on a chemical level [51]. We know the mature virions are small around 100nm in diameter and these are suspended in droplets an order of magnitude larger in size. Exposure to these can be reduced by surgical mask and stopped by N95 masks [52].
We know the key mechanism of molecular biological infection is centred around the SARS-CoV-2 Spike protein. This means mutations on this protein paticularly around the receptor-binding domain will affect infectivity – this allows new dangerous strains to be quickly identified.
Vaccination is the major tool in tackling the pandemic, but in time it is likely new variants will become increasingly resistant to natural and vaccine acquired immunity – work on updated vaccines is already underway [53]. Only with a full set of tools, from vaccines to tissues, mask to hand sanitiser, will the virus be overcome. With such a slow worldwide vaccination program. Travel restrictions may be especially important to stop variants toing and froing between resistant and vulnerable populations. Although, the worst may be behind us for countries like the UK, much remains to be done for poorer nations. We are no longer playing catch-up with Covid-19 scientifically speaking, but only with education and understanding will we be able to keep pace.
References:
1 – Liu, YC, Kuo, RL, Shih, SR 2020, “COVID-19: The first documented coronavirus pandemic in history.”, Biomedical Journal, vol. 43, no. 4, pp. 328-333, DOI: https://doi.org/10.1016/j.bj.2020.04.007.
2 – Maragakis, LL, 2020, Coronavirus and COVID-19: Younger Adults Are at Risk Too, Johns Hopkins Medicine, viewed 18th June 2021 <https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/coronavirus-and-covid-19-younger-adults-are-at-risk-too>
3 – WHO 2021, WHO Coronavirus (Covid-19) Dashboard, WHO, Viewed 18th June 2021 < https://libraryguides.vu.edu.au/harvard/internet-websites>
4 – Benvenuto, D et al. 2020, “Evidence for mutations in SARS-CoV-2 Italian isolates potentially affecting virus transmission.”, Journal of Medical Virology, vol. 92, no. 10, pp. 2232-2237, DOI:10.1002/jmv.26104
5 – Wells CR, Sah, P, Seyed, et al. 2020, “Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak”, PNAS, vol. 117, no. 13, pp. 7504-7509, DOI: 10.1073/pnas.2002616117
6 – Slater, J, Masih, N, Irfan, S, 2021, Coronavirus has crushed India’s health system. Patients are on their own., The Washington Post, Viewed 19th of June 2021 <https://www.washingtonpost.com/world/2021/04/27/india-coronavirus-health-care/>
7 – Garg, S, Kim L, Whitaker, M, et al. 2020, “Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019 — COVID-NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep” CDC, vol 69, pp. 458–464. DOI: http://dx.doi.org/10.15585/mmwr.mm6915e3external icon.
8 – Dashti, H, Roche, EC, Bates, DW, et al. 2020, “SARS2 simplified scores to estimate risk of hospitalization and death among patients with COVID-19.”, Sci Rep, vol. 11, no. 4945, DOI https://doi.org/10.1038/s41598-021-84603-0
9 – Shah ASV, Wood R, Gribben C, et al. 2020, “Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study.” BMJ, vol. 371, no. m3582, DOI:10.1136/bmj.m3582
10 – Ritchie, H, Ortiz-Ospina, E, Beltekian, D et al. 2020, Mortality Risk of COVID-19, Our Wold Data, viewed 19th of June <https://ourworldindata.org/mortality-risk-covid#the-current-case-fatality-rate-of-covid-19>
11 – Wikipedia 2021, Basic reproduction number, Wikipeadia, viewed 29th of June 2021 <https://en.wikipedia.org/wiki/Basic_reproduction_number>
12 – UK Government 2021, Vaccination in the United Kingdom, GOV.UK, viewed 29th of June 2021 <https://coronavirus.data.gov.uk/details/vaccinations>
13 – Mahase, E. 2021, “Delta variant: What is happening with transmission, hospital admissions, and restrictions?”, BMJ, vol. 373, no. 1513, DOI:10.1136/bmj.n1513
14 – CDC 2021, Scientific Brief: SARS-CoV-2 Transmission, CDC, Viewed 19th of June 2021 < https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/sars-cov-2-transmission.html>
15 – Zayas, G., Chiang, M.C., Wong, E. et al. 2021, “Cough aerosol in healthy participants: fundamental knowledge to optimize droplet-spread infectious respiratory disease management.”, BMC Pulm. Med., vol. 12, no. 11, DOI:https://doi.org/10.1186/1471-2466-12-11
16 – Atkinson J, Chartier Y, Pessoa-Silva CL, et al., editors. Natural Ventilation for Infection Control in Health-Care Settings. Geneva: World Health Organization; 2009. Annex C, Respiratory droplets. Available from: https://www.ncbi.nlm.nih.gov/books/NBK143281/
17 – Hammett, E. 2020, “How long does Coronavirus survive on different surfaces?”, BDJ Team, vol. 7, pp. 14–15, https://doi.org/10.1038/s41407-020-0313-1
18 – Mason, RJ, 2020, “Pathogenesis of COVID-19 from a cell biology perspective.”, European Respiratory Journal, vol. 55, no. 4, pp. 2000607, DOI: 10.1183/13993003.00607-2020
19 – Hoffmann, M, Kleine-Weber, H, Schroeder, S, et al. 2020, “SARS-CoV-2 Cell Entry Depends on ACE2and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor.’, Cell, vol. 181, pp. 271–280, DOI: https://www.cell.com/cell/pdf/S0092-8674(20)30229-4.pdf
20 – V’kovski, P, Kratzel, A, Steiner, S, et al. 2021, “Coronavirus biology and replication: implications for SARS-CoV-2.”, Nat Rev Microbiol, vol. 19, pp. 155–170, DOI:https://doi.org/10.1038/s41579-020-00468-6
21 – Saraste, J, Prydz, K, 2021, “Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space.”, Cells, 2021, vol. 10, no. 503, DOI: https://doi.org/10.3390/cells10030503
22 – Ke, Z, Oton, J, Qu, K et al. 2020, ‘Structures and distributions of SARS-CoV-2 spike proteins on intact virions.’, Nature, vol. 588, pp. 498–502, https://doi.org/10.1038/s41586-020-2665-2.
23 – Varga, Z, Flammer, AJ, Steiger, P, Haberecker, M, et al. 2020, ‘Electron microscopy of SARS-CoV-2: a challenging task – Authors’ reply.’, The Lancet [Correspondence], vol. 395, no. 10238, pp. e100, DOI:https://doi.org/10.1016/S0140-6736(20)31185-5.
24 – Meer GV 1998, ‘Lipids of the Golgi membrane’, trends in Cell Biology, vol. 8.
25 – Heo L and AlphaFold colleagues, SARS-Cov-2 protein structure models, GitHub, viewed 21 April 2021, <https://github.com/feiglab/sars-cov-2-proteins>.
26 – Katsnelson A C&EN 2020, What do we know about the novel coronavirus’s 29 proteins?, Chemical and Engineering News, Viewed 19th of June 2021 <https://cen.acs.org/biological-chemistry/infectious-disease/know-novel-coronaviruss-29-proteins/98/web/2020/04>
27 – Dongwan, K, Joo-Yeon, L, Jeong-Sun, Y, Jun, WK, Narry, KV, Hyeshik, C, 2020, ‘The Architecture of SARS-CoV-2 Transcriptome’, Cell, vol. 181, no. 4, pp. 914-921.e10, https://doi.org/10.1016/j.cell.2020.04.011.
28 – Schoeman, D, Fielding, BC, 2019, “Coronavirus envelope protein: current knowledge.”, Virol. J., vol. 16, no. 69, DOI:https://doi.org/10.1186/s12985-019-1182-0
29 – Korber, B, Fischer, WM, Gnanakaran, S, et al. 2021, “Tracking Changes in SARS-CoV-2 Spike: Evidencethat D614G Increases Infectivity of the COVID-19 Virus.”, Cell, vol. 182, pp. 812–827, link: https://www.cell.com/cell/pdf/S0092-8674(20)30820-5.pdf
30 – Liangwei, D, Qianqian, Z, Hongxia, Z, Yuna, N, Yunwei, L, Hui, W, 2020, ‘The SARS-CoV-2 Spike Glycoprotein Biosynthesis, Structure, Function, and Antigenicity: Implications for the Design of Spike-Based Vaccine Immunogens.’, Frontiers in Immunology, vol. 11, pp. 2593, DOI: https://www.frontiersin.org/article/10.3389/fimmu.2020.576622
31 – Public Health England 2020, Investigation of novel SARS-COV-2 variant – Variant of Concern 2020/12/01, GOV.UK, viewed 29 of June 2021 < https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf >
32 – Wikipeadia 2021, SARS-CoV-2 Alpha variant, Wikipeadia, viewed 20th of June 2021 <https://en.wikipedia.org/wiki/SARS-CoV-2_Alpha_variant>
33 – Hart, R 2021, Delta Variant Fueling U.K. Covid Surge Could Become Dominant U.S. Strain. Here Are The Places Most At Risk., Forbes, viewed on the 20th June 2021 <https://www.forbes.com/sites/roberthart/2021/06/14/delta-variant-fueling-uk-covid-surge-could-become-dominant-us-strain-here-are-the-places-most-at-risk/?sh=1a4c391e4e31>
34 – Wikipeadia 2021, SARS-CoV-2 Delta variant, Wikipeadia, viewed on the 20th of June 2021 <https://en.wikipedia.org/wiki/SARS-CoV-2_Delta_variant>
35 – Wikipedaia 2021, SARS-CoV-2 Beta variant, Wikipeadia, viewed on the 20th of June 2021 <https://en.wikipedia.org/wiki/SARS-CoV-2_Beta_variant>
36 – Walls, AC, Park, JY, Tortorici, MA, Wall, A, et al. 2020, ‘Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.’, Cell, vol. 181, no. 2, pp. 281-292.e6, https://doi.org/10.1016/j.cell.2020.02.058.
37 – Xie, Y, Karki, CB, Dan, Du, et al, 2020, ‘Spike Proteins of SARS-CoV and SARS-CoV-2 Utilize Different Mechanisms to Bind With Human ACE2.’, Frontiers in Molecular Biosciences, vol. 7, pp. 392, DOI: https://www.frontiersin.org/article/10.3389/fmolb.2020.591873
38 – Cai, Y, Zhang, J, Xiao, T, Peng, H, et al. 2020, ‘Distinct conformational states of SARS-CoV-2 spike protein.’, Science, pp. 1586-1592.
39 – Yao, H, Song, Y, Chen, Y, Wu, N, et al. 2020, ‘Molecular Architecture of the SARS-CoV-2 Virus.’, Cell, vol. 183, no. 3, pp. 730-738.e13, https://doi.org/10.1016/j.cell.2020.09.018.
40 – Neuman, BW, Adair, BD, Yoshioka, C, Quispe, JD, et al. 2006, ‘Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy.’, J. Virol., vol. 80, no. 16, pp. 7918-28, doi: 10.1128/JVI.00645-06.
41 – Sunil, T, 2020, “The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET.”, Pathogens & immunity, vol. 5, no. 1, pp. 342-363, DOI:10.20411/pai.v5i1.377
42 – Mandala, VS, McKay, MJ, Shcherbakov, AA et al. 2020, ‘Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.’, Nat. Struct. Mol. Biol., vol. 27, pp. 1202–1208, https://doi.org/10.1038/s41594-020-00536-8.
43 – Andrade, MA, Bianchi, M, Benvenuto, D, Giovanetti, M et al. 2020, ‘Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics?’, BioMed Research International, vol. 2020, no. 4389089, https://doi.org/10.1155/2020/4389089.
44 – NHS 2021, Coronavirus (COVID-19) vaccines, uk.gov, viewed 20th of June 2021 <https://www.nhs.uk/conditions/coronavirus-covid-19/coronavirus-vaccination/coronavirus-vaccine/>
45 – Corum, J, Zimmer, J 2021, How the Oxford-AstraZeneca Vaccine Works., The New York Times, viewed 20th of June 2021 <https://www.nytimes.com/interactive/2020/health/oxford-astrazeneca-covid-19-vaccine.html>
46 – Pfizer 2021, Facts About the Pfizer-BioNTech Covid-19 Vaccine, Pfizer, viewed on the 20th of June 2021 <https://www.pfizer.com/news/hot-topics/the_facts_about_pfizer_and_biontech_s_covid_19_vaccine>
47 – Gallagher J 2020, Oxford vaccine: How did they make it so quickly?, BBC News, viewed 20th of June 2021 <https://www.bbc.co.uk/news/health-55041371>
48 – Wellcome 2021, How have Covid-19 vaccines been made quickly and safely?, Wellcome, viewed 20th of June 2021 <https://wellcome.org/news/quick-safe-covid-vaccine-development>
49 – WHO 2021, The Pfizer BioNTech COVID-19 vaccine: What you need to know., WHO, viewed on the 20th of June 2021 <https://www.who.int/news-room/feature-stories/detail/who-can-take-the-pfizer-biontech-covid-19–vaccine>
50 – WHO 2021, The Oxford/AstraZeneca COVID-19 vaccine: what you need to know, WHO, viewed on the 20th of June 2021 <https://www.who.int/news-room/feature-stories/detail/the-oxford-astrazeneca-covid-19-vaccine-what-you-need-to-know>
51 – UNESCO 2020, How Soap Kills COVID-19 on Hands, UNESCO, viewed 20th of June 2021 <https://en.unesco.org/news/how-soap-kills-covid-19-hands>
52 – Tcharkhtchi, A, Abbasnezhad, N, Zarbini, M, 2021, “Overview of filtration efficiency through the masks: Mechanisms of the aerosols penetration.”, Bioactive Materials, vol. 6, no. 1, pp. 106-122, DOI: https://doi.org/10.1016/j.bioactmat.2020.08.002.
53 – Callaway, E, Ledford H, 2021, “How to redesign COVID vaccines so they protect against variants.”, Nature News, vol. 590, pp. 15-16, link: https://www.nature.com/articles/d41586-021-00241-6